How to measure plasma and mitochondrial membrane potential in an unbiased manner
The assay comprises recording and analysis parts. You perform the recording on an arbitrary live-cell fluorescence microscope setup based on our protocols and publications. You use your own reagents or our reagents that will be available later. You perform image and data analysis in Image Analyst MKII. Purchased software license allows all analysis required to perform the analysis.
- Adherent or immobilized cell cultures in glass-bottomed microplates are amenable to the assay.
- To keep conditions as physiological as possible, we designed specialty assay media based on nonfluorescent culture media, where glucose, pyruvate, glutamine, and Ca2+ can be varied.
- A fluorescence time course is recorded in two channels, looking at tetramethylrhodamine methyl ester (TMRM) and FLIPR membrane potential probe (aka. PMPI) fluorescence using an arbitrary motorized microscope.
- During recording, cultures may be challenged multiple times by part-replacement of the potentiometric assay medium.
- The arbitrary time course recording is followed by a standardized
calibration paradigm. This allows calculation of millivolt values for
the entire time course by Image Analyst MKII for both ΔψP and ΔψM. Calibration
points for “complete calibration”:
- zero ΔψM
- stepwise increments of [K+]EC to depolarize ΔψP
- zero ΔψP
- Two cell type-specific geometric parameters are measured in separate experiments using confocal microscopy (or looked up from tables).
- Wet-bench assay protocols are available here.
- Image and data analysis is aided by an interactive protocol in Image Analyst Primer Window (see below).
The Potentiometric Assay - Experimental design and data analysis
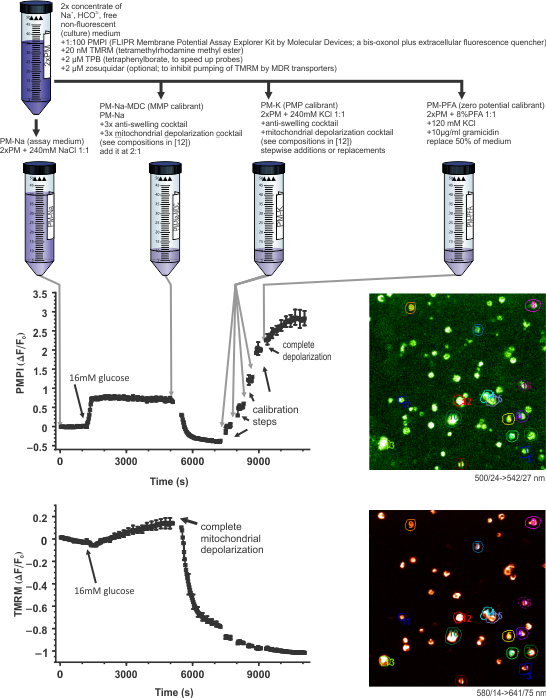
Step 1: The potentiometric assay paradigm
The assay can be performed on any epifluorescence (suitable for low-light level time-lapse imaging), confocal or two-photon system. A prototype experiment is exemplified below using human pancreatic beta-cells (published in (26)). An arbitrary time course recording is followed by a standardized calibration paradigm. The calibrant cocktails have been described in (17), and their preparation in (61).
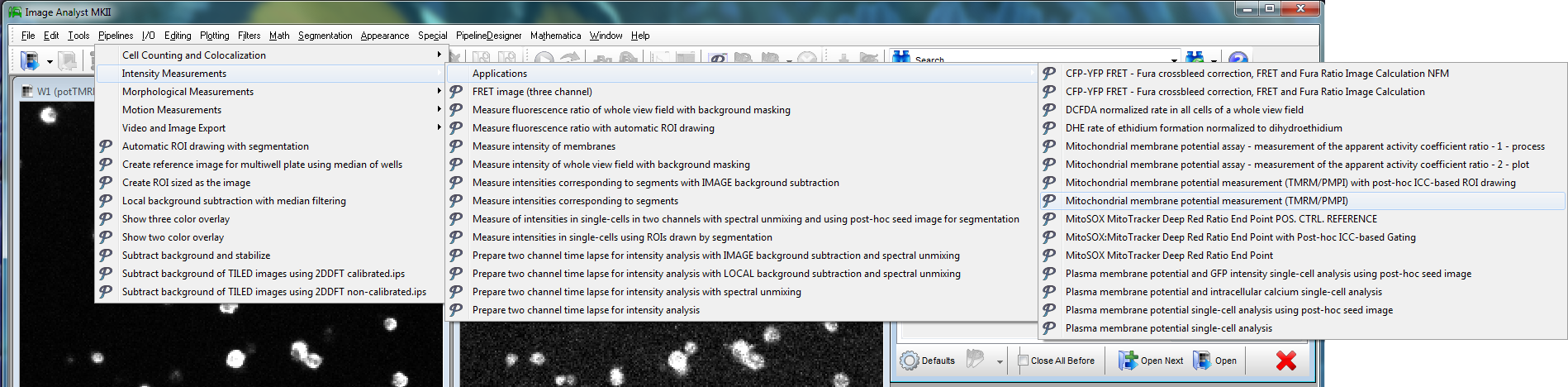
Step 2: Image Analysis
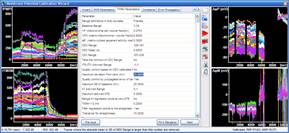
The mitochondrial membrane potential analysis can now be performed using an interactive protocol in the Primer Window by selecting Assays/Intensity and Ratio Measurements/ Mitochondrial membrane potential assay - worked example. Buttons in the protocol actually operate Image Analyst MKII.
Alternatively, select from a set of pre-configured pipelines in the main menu of Image Analyst MKII to process fluorescence time-lapse image recordings and automatically draw ROIs to extract fluorescence intensities. See tutorial image data for potentiometric image analysis here, and a video tutorial here.
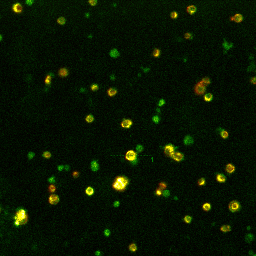
Left: Microscopic
view-field of TMRM (red) and FLIPR (green) fluorescence in
pancreatic beta-cells.
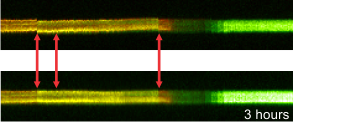
Right: Effect of channel alignment
and image stabilization of the time lapse, demonstrated by a
line scan
These image processing steps are included into the standard pre-processing pipelines for potentiometric calibrations.
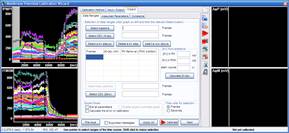
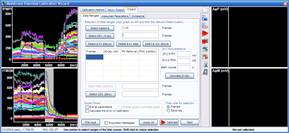
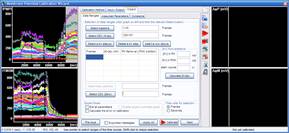
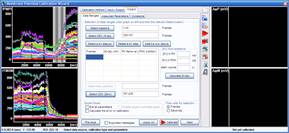
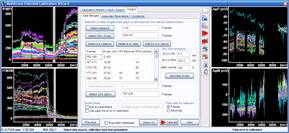
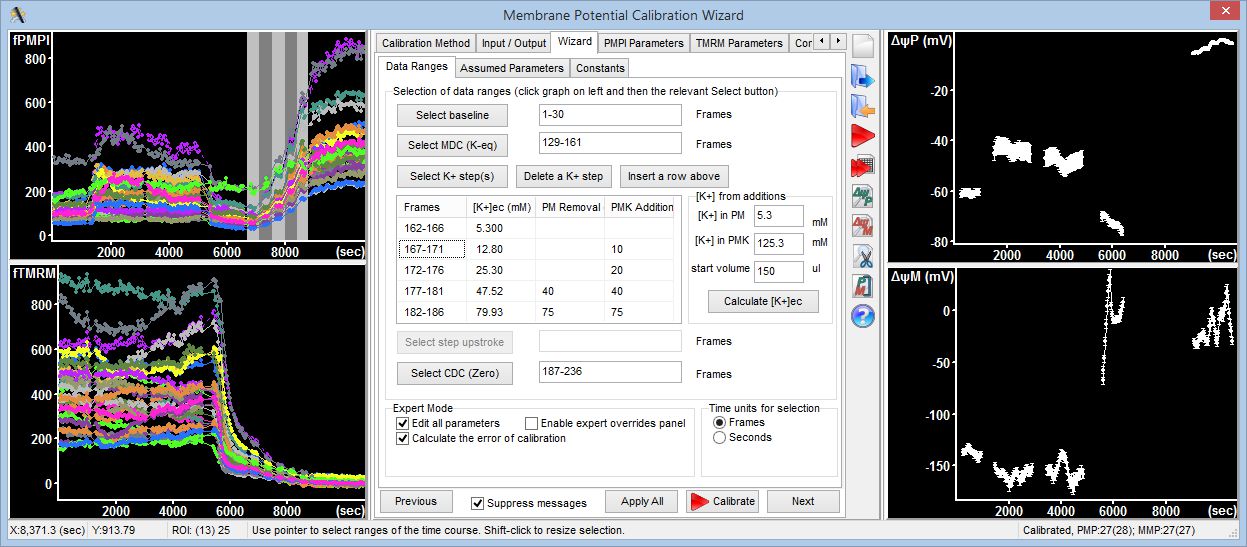
Step 3: Calculation of millivolts using the Membrane Potential Calibration Wizard
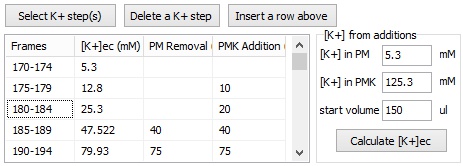
The Membrane Potential Calibration Wizard guides the user through the calibration procedure. The user selects the experimental paradigm matching the recording, then points the ranges of calibrant additions on the fluorescence time courses, and finally enters required additional parameters. In the complete calibration paradigm only volumes of K+-based medium additions are required to perform the calibration.
- Complete (temporally resolved K-steps)
- Complete with known kP (K-steps)
- Baseline & Zero
- Baseline & K-equilibrium
- K-equilibrium & Zero
- K-steps with known [K+]i
- Zero (fx=0)
- Baseline & fP0
- Complete
- Complete (known k)
- Baseline & MDC or CDC (known rest)
Step 4: Automation
Using pipeline automation the Membrane Potential Calibration Wizard can be executed completely automatically in each stage position or well of a microplate, analyzing thousands of individual cells in a single run. Millivolt calibrated data can be collected in Graphpad Prism from multiple conditions and experiments including pooling technical replicates.
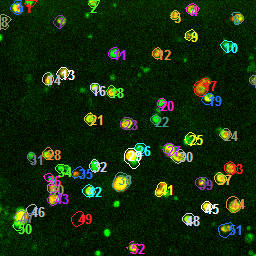
As part of the automated pipeline processing, individual cells are identified by an automatic ROI drawing feature.
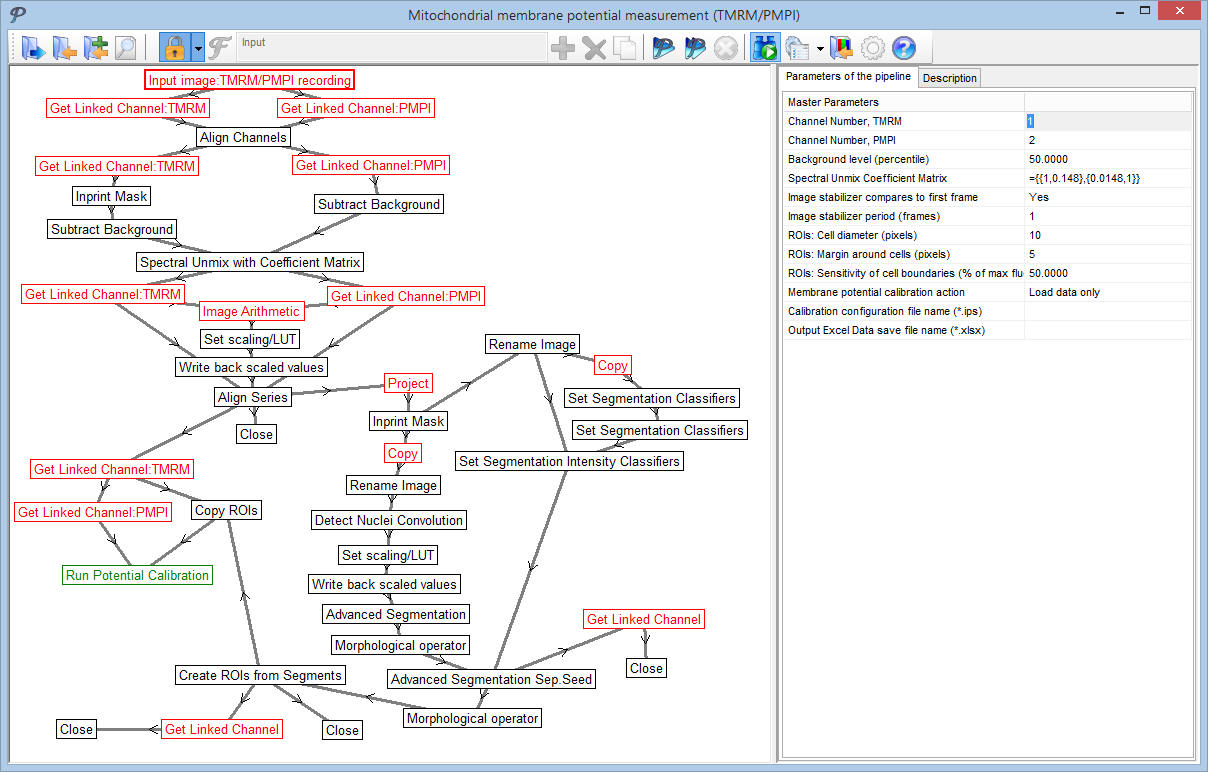
The menu-accessible pipelines are invisible for normal operation, and only key-parameters are shown in the main parameter bar. However, pipelines can be opened for editing and arbitrarily changed.
| Development of the unbiased, absolute mitochondrial membrane potential assay has been supported by: |

 |
Theory Overview